Research
化学を基盤として細胞内現象を解析・制御するための新規分子デザインや新手法を考案し、生命科学研究・創薬研究を展開します。
Development of mRNA medicines
-
PureCap method
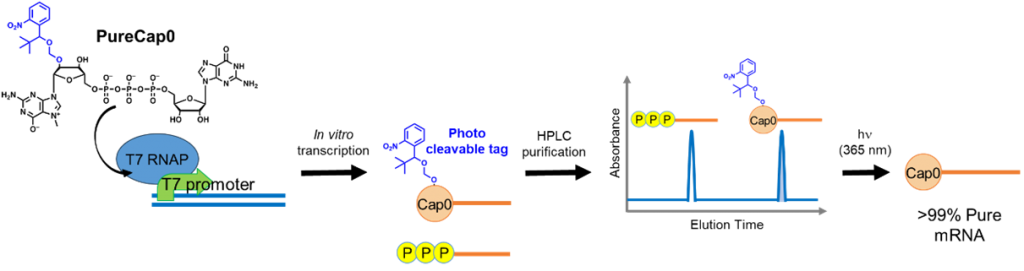
We developed a novel Cap analog to synthesize and purify mRNA with a 5′ Cap structure. The Cap analog, PureCap0, can introduce the Cap0 structure and hydrophobic nitrobenzyl modification as a purification tag on mRNA by in vitro co-transcription. HPLC purification following the transcription can remove the uncapped byproduct altogether. Subsequent photoirradiation at 365 nm removes the purification tag and generates 100% pure capped mRNA. We named this the PureCap method and paved the way for the development of mRNA therapeutics. We have also developed other PureCap analogs introducing Cap1 or Cap2 for mRNA.
References
-
- Inagaki, M., Abe, N., Li, Z. et al. Cap analogs with a hydrophobic
photocleavable tag enable facile purification of fully capped mRNA with various cap structures.
Nat. Commun. 14, 2657 (2023).
https://doi.org/10.1038/s41467-023-38244-8
- Inagaki, M., Abe, N., Li, Z. et al. Cap analogs with a hydrophobic
-
-
Rolling circle translation using circular mRNA
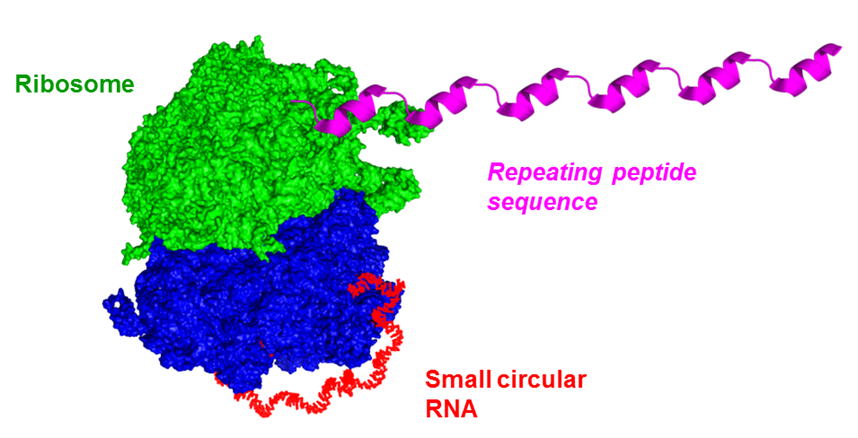
環状RNAを用いた終わりのない回転式タンパク質翻訳 Unlike conventional translation that starts from the initiation codon and terminates at the stop codon, we have demonstrated that circular mRNA, which lacks the stop codon, can perform continuous translation. This novel approach has shown a dramatic increase in protein production, potentially revolutionizing the field of molecular biology.
References
-
- Abe, N., Hiroshima, M., Maruyama, H., et al. Rolling circle amplification in a prokaryotic translation system using small circular RNA, Angewandte Chemie International Edition52(27), 7004-7008 (2013).
https://doi.org/10.1002/anie.201302044 - 環状mRNAを用いてエンドレスなタンパク質合成に成功
https://www.riken.jp/press/2013/20130522_1/
- Abe, N., Hiroshima, M., Maruyama, H., et al. Rolling circle amplification in a prokaryotic translation system using small circular RNA, Angewandte Chemie International Edition52(27), 7004-7008 (2013).
-
-
Complete chemical synthesis of mRNA
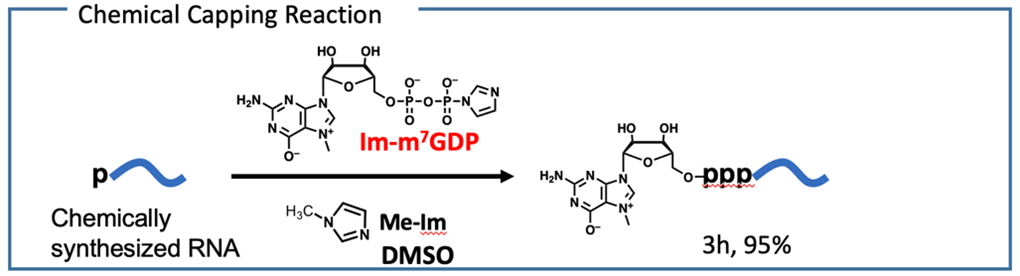
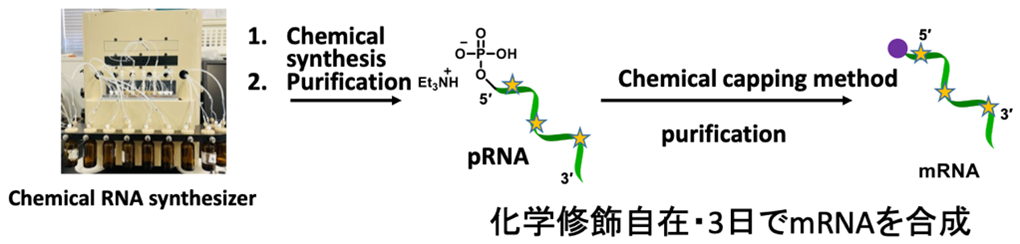
We have conducted the complete chemical synthesis of mRNA. It requires the introduction of 5′ cap structure by chemical scheme. To achieve this, we have developed a chemical capping reaction that quantitatively introduces a cap structure to mRNA in an organic solvent. The chemical synthesis enables us to prepare mRNA, which contains modifications at arbitrary positions to enhance stability and translation efficacy. It is also expected that chemical synthesis can produce mRNA in the short term compared to enzymatic transcription.
References
-
- Abe, N., Imaeda, A., Inagaki, M. et al. Complete chemical synthesis of minimal messenger RNA
by efficient chemical capping reaction. ACS Chem. Biol. 17(6), 1308–1314 (2022).
https://doi.org/10.1021/acschembio.1c00996
- Abe, N., Imaeda, A., Inagaki, M. et al. Complete chemical synthesis of minimal messenger RNA
-
Drug delivery system(DDS)
-
MPON
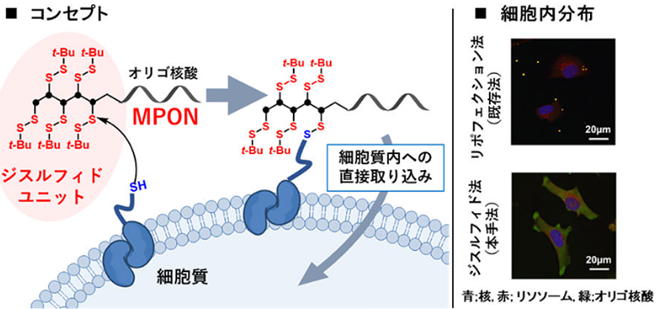
Although nucleic acid drugs, including siRNA and antisense nucleotide, are functional in the cytosol, their membrane permeabilities are quite low because of negative charge. Therefore, delivery methods of nucleic acid molecules for cytosol are attractive topics. We have developed a novel delivery method named membrane permeable oligonucleotide (MPON). MPON contains disulfide units on the nucleotide end, which interact with membrane proteins on the cell surface to enhance membrane permeability. We have demonstrated that siRNA or antisense nucleotide modified with MPON showed more significant gene silencing effects than conventional lipofection for cultured cells. Furthermore, we have also achieved efficient mRNA delivery using the lipid nanoparticle (LNP) in which the cationic lipids are modified with the disulfide units. This scheme applies not only to the delivery of nucleic acid drugs but also to other biomolecules like antibodies or peptides, paving the way for new molecular target drugs.
References
-
- Shu Z, Tanaka I, Ota A, Fushihara D, Abe N, Kawaguchi S, Nakamoto K, Tomoike F, Tada S, Ito Y, Kimura Y, Abe H. Disulfide-Unit Conjugation Enables Ultrafast Cytosolic Internalization of Antisense DNA and siRNA. Angew Chem Int Ed Engl. 2019 May 13;58(20):6611-6615. doi: 10.1002/anie.201900993. Epub 2019 Apr 5. PMID: 30884043.
Shu Z, Ota A, Takayama Y, Katsurada Y, Kusamori K, Abe N, Nakamoto K, Tomoike F, Tada S, Ito Y, Nishikawa M, Kimura Y, Abe H. Intracellular Delivery of Antisense DNA and siRNA with Amino Groups Masked with Disulfide Units. Chem Pharm Bull (Tokyo). 2020;68(2):129-132. doi: 10.1248/cpb.c19-00811. PMID: 32009079. - Hiraoka H, Shu Z, Tri Le B, Masuda K, Nakamoto K, Fangjie L, Abe N, Hashiya F, Kimura Y, Shimizu Y, Veedu RN, Abe H. Antisense Oligonucleotide Modified with Disulfide Units Induces Efficient Exon Skipping in mdx Myotubes through Enhanced Membrane Permeability and Nucleus Internalization. Chembiochem. 2021 Dec 10;22(24):3437-3442. doi: 10.1002/cbic.202100413. Epub 2021 Oct 22. PMID: 34636471.
- Shu Z, Tanaka I, Ota A, Fushihara D, Abe N, Kawaguchi S, Nakamoto K, Tomoike F, Tada S, Ito Y, Kimura Y, Abe H. Disulfide-Unit Conjugation Enables Ultrafast Cytosolic Internalization of Antisense DNA and siRNA. Angew Chem Int Ed Engl. 2019 May 13;58(20):6611-6615. doi: 10.1002/anie.201900993. Epub 2019 Apr 5. PMID: 30884043.
-
-
Lipid nanoparticle(LNP)
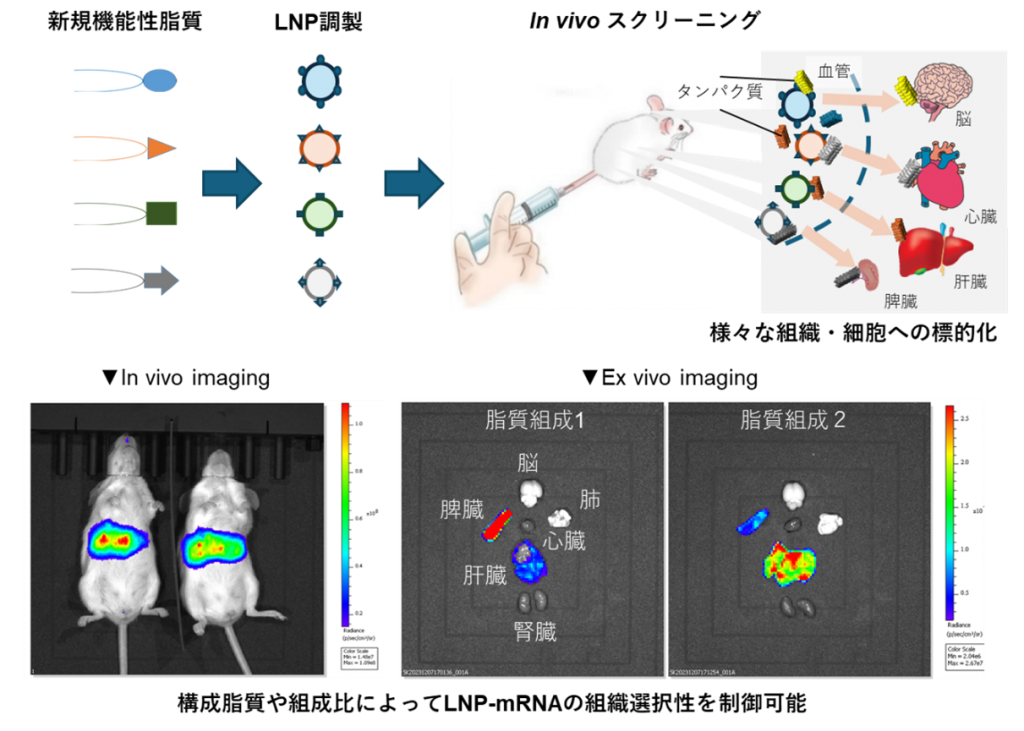
脂質ナノ粒子(LNP)は、siRNA医薬品のオンパットロ、COVID-19に対するmRNAワクチンとして実用化された薬物送達技術であり、今後もその技術応用が加速していくと考えられる。しかし、投与部位の炎症、肝臓外組織への標的化、長期保存安定性等の課題があり、さらなる技術的進展が望まれている。LNPの体内動態・細胞内動態といった機能的特性は、構成脂質の化学構造及び組成比によって規定される。我々は、脳や心臓など、これまで導入が困難であった組織や細胞へ安全かつ効率的に送達可能なLNP開発を目的に、新規機能性脂質の設計・合成及びそれらの機能評価をin vitro(細胞実験)/in vivo(動物実験)で進めている。
References
-
- Kimura S, Harashima H. Nano-Bio Interactions: Exploring the Biological Behavior and the Fate of Lipid-Based Gene Delivery Systems. BioDrugs. 2024 Feb 12. doi: 10.1007/s40259-024-00647-4. Epub ahead of print. PMID: 38345754.
-
合成生物学
-
ゲノム合成
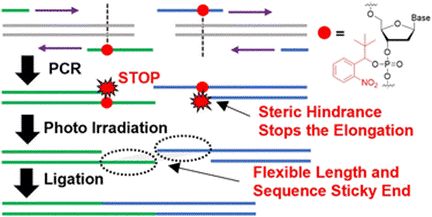
分子生物学の分野ではDNAの操作技術は必要不可欠であり、これを活用したDNAライブラリ構築や遺伝子のクローニングが行われている。mRNA医薬品の作成でも重要であり、特にコロナウイルスのような新興感染症のmRNAワクチン作成では配列情報から速やかにDNA鋳型を作成する必要がある。一般的にはPolymerase Chain reaction (PCR)によって短いDNA断片を作成し、酵素処理によって連結するが、目的のDNAが長くなるにつれて効率が著しく低下し、ゲノムスケールのDNA合成は困難である。我々はstopプライマーと呼ばれる化学合成プライマーを開発し、効率的なDNA連結を達成した。これを活用することで従来法では不可能であった1万塩基の長さを持つDNA断片を複数連結し、大腸菌に感染するウイルスであるラムダファージのゲノムDNA構築に成功した。これを利用して迅速なmRNA転写鋳型やライブラリの構築を可能とする技術開発を行っている。
References
-
- Nomura K, Onda K, Murase H, Hashiya F, Ono Y, Terai G, Oka N, Asai K, Suzuki D, Takahashi N, Hiraoka H, Inagaki M, Kimura Y, Shimizu Y, Abe N, Abe H. Development of PCR primers enabling the design of flexible sticky ends for efficient concatenation of long DNA fragments. RSC Chemical Biology. 2024 Feb 26. Doi: 10.1039/D3CB00212H。
-
